
Peter Kind and colleagues show that brief treatment with lovastatin results in sustained correction of physiological and behavioral deficits in a rat model of fragile X syndrome.
Highlights of SFARI-funded papers, selected by the SFARI science team.

Peter Kind and colleagues show that brief treatment with lovastatin results in sustained correction of physiological and behavioral deficits in a rat model of fragile X syndrome.

Using single-nucleus RNA sequencing of postmortem cortical tissue from individuals with ASD, Arnold Kriegstein and colleagues identify upper-layer excitatory neurons and microglia as key cell types affected in ASD.
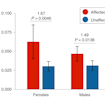
Timothy Yu and colleagues analyzed exome sequencing data to estimate that recessive mutations contribute to approximately 5 percent of all cases of autism, including 10 percent of females.
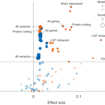
Olga Troyanskaya, Robert Darnell and colleagues applied deep-learning methods to whole-genome sequencing data from SSC families and identified a clear enrichment for de novo noncoding variation in ASD.
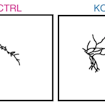
James Ellis and colleagues used a sparse co-culture system for iPSC-derived cortical neurons to assess neuronal connectivity, demonstrating increased connectivity in SHANK2-mediated ASD.

Mark Zylka and colleagues generated single-cell RNA-seq data from wild-type mouse cortex during early development and demonstrated how such a resource can be used to identify putative brain disorder subtypes based on expression profiles.
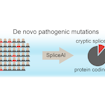
Kyle Kai-How Farh and Stephan Sanders developed a deep-learning method to predict risk mutations that affect mRNA splicing and contribute substantially to neurodevelopmental disorders.
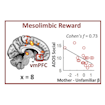
Vinod Menon and colleagues report that a difference in the activation of a voice-processing network comprising reward and salience detection systems is a distinguishing feature of autism.