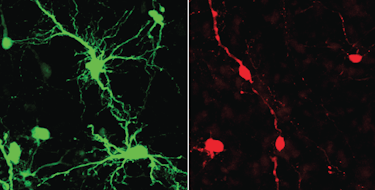
One possible explanation for the differential expressivity of autism spectrum disorder (ASD)-associated phenotypes is allele-biased gene expression. That is, an allele carrying a mutation may have a greater or lesser effect depending on the relative level of expression of that allele. Despite this theoretical possibility, there are few documented examples of allelic imbalance in ASD.
A recent study by SFARI Investigators Daniel Geschwind and Michael Gandal, and their colleagues, sheds new light on the impact of allelic imbalance in ASD. Using a set of postmortem brain tissue samples from frontal cortex and cerebellum, the authors showed that allelic imbalance in gene expression occurs less often in ASD, but when it does occur, it exhibits preferential expression of the minor allele. In other words, the allele carrying the rare mutation shows higher expression in ASD more often than is the case for neurotypical controls. This phenomenon would add to the vulnerability of the ASD brain, since a greater proportion of the gene expression at a given locus would be affected by the mutation. Indeed, loci with allelic imbalance of gene expression are enriched for genes that are known to harbor de novo ASD risk variants.
One of the genomic regions showing a very strong shift to predominant expression of the minor allele is at the ASD-associated 15q11-q13 duplication interval (dup15q syndrome). In particular, their analysis highlights the orphan C/D box small nucleolar RNA (snoRNA) genes in this region, which preferentially target alternatively spliced junctions of downstream ASD risk genes. These results implicate a common set of alternatively spliced targets of snoRNAs in both idiopathic ASD and dup15q-associated ASD and suggest that a careful examination of allelic imbalance may help to explain some of the phenotypic variability in individuals carrying mutations in the same risk gene.

Reference(s)
Profiling allele-specific gene expression in brains from individuals with autism spectrum disorder reveals preferential minor allele usage.
Lee C., Kang E.Y., Gandal M., Eskin E., Geschwind D.