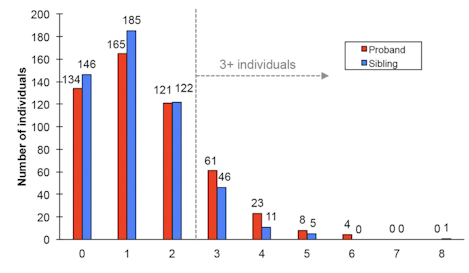
While the autism research community has made great strides in uncovering numerous genetic risk factors for autism spectrum disorder (ASD), there is still much to understand about the genetic risks that arise from spontaneous de novo mutations. To address this issue, SFARI Investigators Evan Eichler and Robert Darnell performed deep whole genome sequencing of 516 Simons Simplex Collection (SSC) families with idiopathic ASD, with one child affected and at least one unaffected sibling. This work has led to the generation of a new database of de novo genetic variants from 2,064 individuals, representing a 10-fold increase in sample size over previously existing SSC data sets. Analysis of this data set confirms that increases in paternal age are associated with an increased risk of de novo substitutions. Further, by focusing on idiopathic ASD families, the authors were able to dissect out how the relative impacts of particular de novo mutations relate to their positioning within noncoding functional elements of the genome. Their analyses suggest that affected individuals are more likely to carry severe, gene-disruptive mutations and that affected individuals often carry multiple de novo mutations in different genes. Notably, this ‘oligogenic’ burden highlights genes whose expression is enriched in D1+ and D2+ spiny neurons in the striatum. These findings advance our understanding of ASD, underscoring the importance of large-scale whole genome sequencing to help elucidate the etiology of simplex ASD.
Reference(s)
Genomic patterns of de novo mutation in simplex autism.
Turner T., Coe B.P., Dickel D.E., Hoekzema K., Nelson B.J., Zody M.C., Kronenberg Z.N., Hormozdiari F., Raja A., Pennacchio L., Darnell R., Eichler E.


