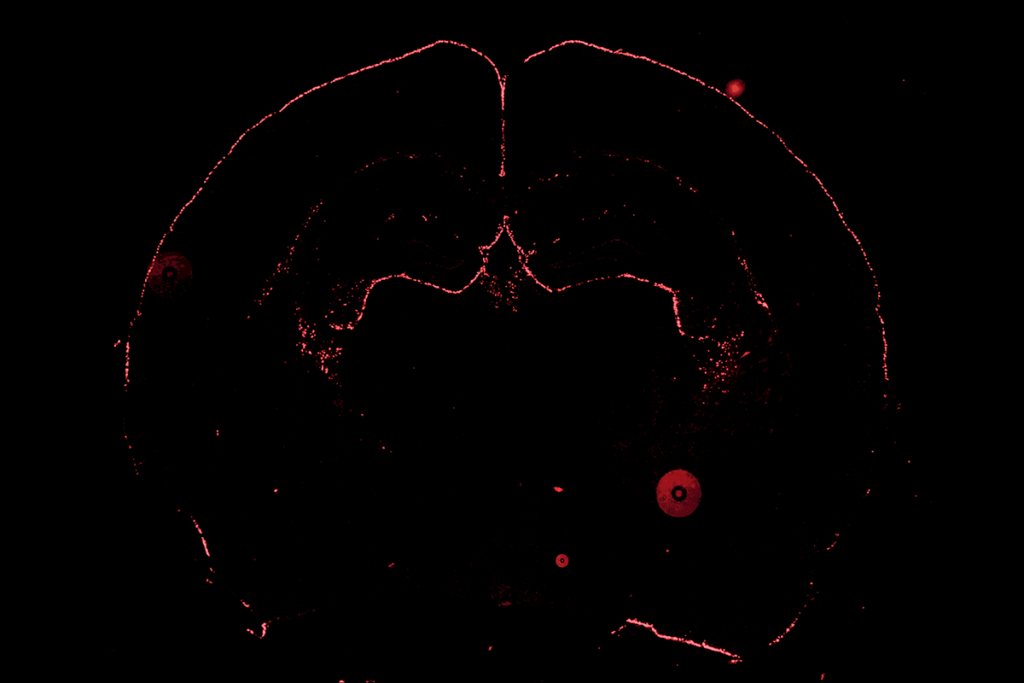
New imaging techniques probe brain’s long-range connections
New techniques to scan the brain can produce exquisitely detailed views of white matter, which contains the long cellular fibers that connect neurons. Many of the advances are emerging from the Human Connectome Project, a five-year push to map the brain’s wiring.
For five years in the late 1990s, Van Wedeen studied the structure of the heart, even though what he really wanted to explore was the wiring of the brain.
Wedeen was biding his time because he knew that existing imaging techniques couldn’t render an accurate picture of the brain’s white matter, which contains the long cellular fibers that connect neurons.
“So there the matter stayed for five or so years, while I turned to other organs and simpler questions,” says Wedeen, assistant professor of radiology at Massachusetts General Hospital.
By the mid-2000s, better scanners and advanced mathematical algorithms allowed Wedeen and others to distinguish tightly packed neuronal wires, or axons, running in different directions. That part of the picture had long been elusive; other techniques show only one direction of travel at a given point in the brain.
Wedeen is using those techniques as he co-leads a team of researchers working on the Human Connectome Project (HCP). Launched in 2010, the project is a five-year effort to describe the structure and function of connections between different regions of the brain in 1,200 healthy adults.
Defining the brain’s wiring diagram can help researchers understand what goes awry in brain disorders such as autism. The popular ‘connectivity theory’ of autism holds that long-range connections are impaired in the brains of people with the disorder.
Wedeen is not alone in pursuing this goal. Another team of HCP investigators has a somewhat different approach, favoring another imaging technique and way of calculating white-matter trajectories.
“It will probably turn out in the end that one method is modestly better than the other, and it will be valuable to sort that out,” says David Van Essen, professor of anatomy and neurobiology at Washington University in St. Louis, a leader of the second team.
Drawing straws:
As part of the $40 million project, both HCP teams have built customized brain scanners, and spent two years refining the methodology for acquiring and processing the images. The first data from the project went online in October.
The results of their methodological debates are likely to have wider significance.
“We’re watching with interest,” says David Edwards, director of the Centre for the Developing Brain at King’s College, London. Edwards is leading the Developing Human Connectome Project, a €15-million European effort to track how brain connectivity develops between 20 and 44 weeks after conception. His team is in close contact with leaders of the HCP, though the two projects are independent.
White matter is tricky to scan because its fibers are only a few micrometers wide, but often ten or more centimeters long.
“You have a three-dimensional (3D) trajectory that you have to be able to trace in the brain over what are fairly long distances,” says Pratik Mukherjee, associate professor of radiology at the University of California, San Francisco. Mukherjee is not involved in either connectome project, but has mapped white matter connections in people lacking a corpus callosum, a birth defect linked to autism.
The only way to trace white matter connectivity — at least, in a living person — is with diffusion imaging, a type of magnetic resonance imaging (MRI) based on the movement, or diffusion, of water inside the brain. White matter fibers act like drinking straws, constraining the movement of water along their length.
“We became aware, 20 years ago or so, that the diffusion signal provided information about the direction of cellular fibers inside the brain,” says Wedeen, who helped develop a technique called diffusion tensor imaging (DTI).
DTI studies have shown differences in particular regions of the brain in people with autism compared with controls1, and in infants at high risk for autism who later develop the disorder compared with those who do not2.
But Wedeen and others say DTI is a relatively crude method that isn’t well suited to mapping the fine-scale detail of the brain’s wiring. That’s because the images carve up the brain into 3D pixels, or voxels — imaginary cubes stacked together to build an image of the brain. In images from standard scanners, these cubes measure a few millimeters or so on a side. And DTI can only indicate one direction of water movement per voxel.
“In many areas of the brain, there are multiple fiber orientations within a voxel,” Mukherjee says. White matter fibers can cross over each other, fan out, converge, or turn sharply. “You cannot get adequate information from DTI in those areas.”
Sharp turns:
Researchers have had to develop new scanning protocols and mathematical models for processing MRI images. Faster scanners now yield higher-resolution images, which means that researchers can divide the brain into smaller voxels. The best resolution comes from state-of-the-art scanners built for the HCP by the engineering and electronics firm Siemens.
But even voxels that measure just over a millimeter per side from these scanners still contain many thousands of nerve fibers running in different directions.
“We have to obtain images where we monitor water molecules diffusing in different directions,” says Kamil Ugurbil, professor of radiology and neuroscience at the University of Minnesota and co-leader of Van Essen’s team.
Two new techniques, diffusion spectrum imaging (DSI) and high angular resolution diffusion imaging (HARDI), accomplish this, each capable of capturing the movement of water in hundreds of directions within every voxel.
These improvements in diffusion imaging are beginning to radically reshape our view of white matter’s architecture.
Last year, Wedeen reported data suggesting that white matter fibers traverse the brain by making a series of sharp 90-degree turns in a 3D grid, rather than in long, swooping arcs as researchers had initially assumed3.
Wedeen says he has unpublished microscopy data validating the existence of these ‘micro-turns.’ Others aren’t yet convinced that his proposal represents a fundamental insight into the structure of white matter.
“This is one where I would say I hope it’s true, but to me the jury’s still out,” says Van Essen. “And if it’s true in some places, it may not be true everywhere.”
Van Essen and Wedeen plan to conduct a side-by-side comparison of their techniques to work out which approach yields more useful results.
Meanwhile, researchers working on the European project have also been refining their methods. They plan to include brain images of fetuses in utero and preterm infants, and have developed mathematical methods to correct for movement during the fetal scans, which would normally wreck an MRI image.
The team plans to scan at least 1,200 participants, beginning in September 2014. The data will be made freely available online and compatible with HCP data as much as possible. “They’re a couple of years ahead of us, so we’ll be taking advantage of what they’ve learned,” Edwards says.
Image courtesy of the Laboratory of Neuro Imaging at UCLA and Martinos Center for Biomedical Imaging at MGH, Consortium of the Human Connectome Project.
References:
1: Lange N. et al. Autism Res. 3, 350-358 (2010) PubMed
2: Wolff J.J. et al. Am. J. Psychiatry 169, 589-600 (2012) PubMed
3: Wedeen V.J. et al. Science 335, 1628-1634 (2012) PubMed
Recommended reading
New tool may help untangle downstream effects of autism-linked genes
NIH neurodevelopmental assessment system now available as iPad app
Molecular changes after MECP2 loss may drive Rett syndrome traits
Explore more from The Transmitter
Who funds your basic neuroscience research? Help The Transmitter compile a list of funding sources

The future of neuroscience research at U.S. minority-serving institutions is in danger