
SFARI initiated whole-genome sequencing (WGS) of the Simons Simplex Collection (SSC) in early 2015. This update is intended to inform the research community about the latest data that are now available, including an additional 6,801 whole genomes as well as reprocessed data from previously released genomes.
Data availability
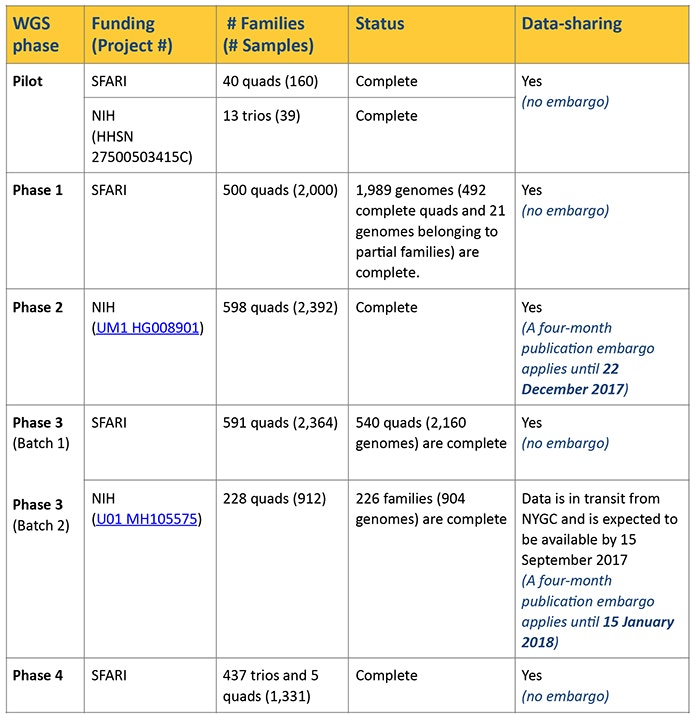
A total of 2,174 whole genomes have been available since August 2016, including most of the samples from the Pilot study and Phase 1 (see Table 1). For these data, alignment of reads to hg19/ National Center for Biotechnology Information (NCBI) build 37 was done using Burrows-Wheeler Aligner (BWA) software package (BWA-MEM), and all local realignment and variant calling was done using Genome Analysis Toolkit (GATK) best practices (version 3.4). These data have now been reprocessed using a different computational pipeline, which uses BWA-MEM (0.7.15) alignment of reads to hg38/NCBI build 38, with adjustments to sorting of duplicate markings and base quality score recalibration. Variant calling was done using GATK (version 3.5).
In addition to this reprocessed dataset, we are also releasing BAM (Binary Alignment/Map), CRAM (compressed versions of the alignment) and VCF (variant call format) files for an additional 6,801 whole genomes (i.e., 1,369 quad families [5,476 genomes], 437 trio families [1,311 genomes] and 14 genomes from partial families). Structural variant calls are being processed and will be made available as soon as possible. These new genomes were sequenced in batches and were processed using the same computational pipeline employed for the reprocessing of the Pilot study and Phase 1 data, as described above. The sequencing batches and publication embargoes (if applicable) are outlined in Table 1.
Note: Most of the data detailed in Table 1 are currently available, although some datasets are still in transit from the New York Genome Center (NYGC) and will be available in the coming weeks. Furthermore, there are an additional 86 probands with incomplete families that will be released in the near future.
Which researchers can use the WGS data?
WGS data are available for use by all approved researchers, regardless of SFARI funding. The following restrictions apply:
1) The intended research must be relevant to autism and/or other neurodevelopmental disorders to comply with consents.
2) Researchers must agree to abide by any publication embargos that apply.
Accessing the data
Researchers can access the data by logging into SFARI Base and completing an application. The application will be reviewed, and once approved, the data can be accessed via the following three options:
- Cloud-based access (via Amazon Web Services)
- Fermilab access (data can be transferred to research institute servers via GridFTP)
- Simons Foundation server (data can be transferred to research institute servers via globus.org)
Additional details about how to download or access the data (estimated to be roughly 400TB for all of the data) will be provided to researchers after their SFARI Base application has been approved.
Additional information
- A PDF version of this announcement is available here.
- A PDF document that outlines Frequently Asked Questions (FAQs), including additional information about sample selection and data access, is available here.
- Comparison of the pipelines developed by the Centers for Common Disease Genomics (CCDG) for hg38 (NCBI build 38) and the NYGC pipeline for hg19 (NCBI build 37) is available here.
- Information on the CCDG compliant pipeline is available in the github repository.
- Additional information about the SSC (including a list of publications that have used SSC genomic data) is available here.


