THIS ARTICLE IS MORE THAN FIVE YEARS OLD
This article is more than five years old. Autism research — and science in general — is constantly evolving, so older articles may contain information or theories that have been reevaluated since their original publication date.
Mutations in FAN1, a gene in the 15q13.3 chromosomal region, raise the risk of neuropsychiatric disorders including autism and schizophrenia, according to a new study published 7 January in the Proceedings of the National Academy of Sciences1.
The 15q13.3 chromosomal region is a hotbed of tiny genetic deletions and duplications connected to disorders of brain development. Which genes within the region are the culprits has been unclear, however.
Prior to this study, researchers’ best guess for the errant gene underlying several disorders was CHRNA7, one of seven genes in the region prone to microdeletions.
“CHRNA7 has been discussed for years now as a prime candidate gene for the psychiatric disturbances associated with these microdeletions,” says Maria Karayiorgou, professor of psychiatry at Columbia University. “So it was quite surprising to find another gene, not previously discussed or suspected, as a major culprit.”
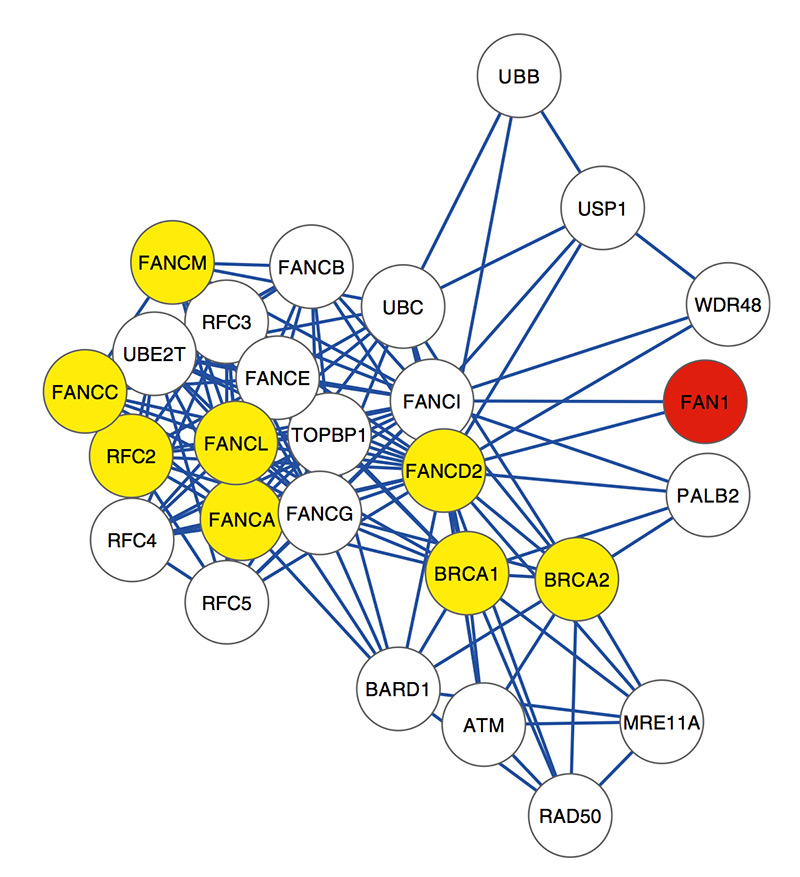
FAN1 is better known for its connection to Fanconi anemia, an inherited blood disorder often accompanied by congenital defects and cancer. It codes for a DNA repair enzyme that helps fix faulty cross-links between DNA strands, which can be deadly for a cell.
“That abnormalities in DNA repair are implicated in the genetic component of schizophrenia and autism is quite surprising and novel,” says Karayiorgou. This raises the intriguing question of whether toxins or other environmental insults that cause DNA damage could cause neurodevelopmental problems in individuals with microdeletions in 15q13.3.
“It’s a very interesting study; it’s very well done,” says Bernard Crespi, professor of evolutionary biology at Simon Fraser University in Burnaby, British Columbia, Canada, who was not involved in the study.
Cluster study:
The researchers hunted down FAN1 using a technique called scan statistic analysis, first used in mathematics in the 1960s and applied to autism in 20012. In that study, the researchers identified small regions of high genetic risk for autism within larger chromosomal regions known to contain rare risk variants.
They assigned a likelihood score for the presence of autism risk variants to subsections of the region. They then scanned the genomes of pairs of siblings, one of whom has autism, recording which stretches have peaks of the highest susceptibility. In these cases, unaffected siblings may possess variants that tend to cause a gene to lose its function, but people without a family history of autism tend not to.
For the new study, Karayiorgou and her colleagues modified the method to home in on clusters of genetic variants within small sections of the exome, or the coding region of the genome. They took advantage of the fact that rare genetic variants tend to cluster together in mutation-rich regions.
They repeatedly screened the same sections of exomes, starting at different points, using a sliding window of 20,000 nucleotides. The technique screens for both association between variants and clusters of them within a region.
In contrast, whole-genome or whole-exome sequencing analysis looks for variants in each individual suspect gene.
“Existing methods don’t take clustering of disease risk variants into account. The method we used does, and I think that is why we were able to detect this association,” says Iuliana Ionita-Laza, assistant professor of statistics at Columbia University in New York.
The researchers analyzed two independent sets of whole-genome sequences from families of individuals with schizophrenia, and one set of exomes of 488 unrelated individuals with autism and 372 matched controls.
FAN1, a single gene on chromosome 15q13.3, stood out, containing clusters of rare mutations that are associated with risk for both autism and schizophrenia.
The researchers also looked for genes that interact with FAN1. They found 27 genes, 10 of which carry rare harmful mutations.
This method of pinpointing a gene is promising, but FAN1 itself may not stand up to further scrutiny as a strong candidate gene for autism and schizophrenia, says Ivan Iossifov, assistant professor of quantitative biology at Cold Spring Harbor Laboratory in New York, who did not participate in the study.
“I think it is a good example for what is to come, in terms of analyzing exome sequencing data on families, particularly if the gene survives,” Iossifov says.
Crespi also cautions that the study is preliminary, has small sample sizes and doesn’t show significant associations between all measures. “But it leads directly to some interesting new questions,” he says, such as why people with autism or schizophrenia have a greater burden of mutations overall.
References:
1: Ionita-Laza I. et al. Proc. Natl. Acad. Sci. USA 1, 343-348 (2014) PubMed
2: Liu, J. et al. Am. J. Hum. Genet. 69, 327–340 (2001) PubMed
By joining the discussion, you agree to our privacy policy.