THIS ARTICLE IS MORE THAN FIVE YEARS OLD
This article is more than five years old. Autism research — and science in general — is constantly evolving, so older articles may contain information or theories that have been reevaluated since their original publication date.
Spontaneous and rare mutations, particularly in genes related to networks that regulate neuronal connections, contribute a small but significant proportion of the risk for schizophrenia, report two large studies published online 22 January in Nature.
One study, led by Michael O’Donovan at Cardiff University in the U.K., found that people with schizophrenia have higher rates of spontaneous mutations in groups of genes involved in synaptic plasticity — the ability of neurons to change the strength of their connections — than controls do1. They also have more mutations in genes that are targets of FMRP, the protein missing in fragile X syndrome. Many of these genes have been linked to autism.
The second study found that a wide variety of rare mutations contribute to schizophrenia and tend to cluster in groups similar to the de novo mutations2. That study was led by Shaun Purcell at the Broad Institute of the Massachusetts Institute of Technology and Harvard University.
| Emily Elert | Sources |
“Both papers demonstrate that there’s extreme genetic heterogeneity in schizophrenia,” says Jon McClellan, professor of psychiatry at the University of Washington, who was not involved in either study.
Each is the largest study to date looking at its particular type of mutation, de novo or rare variants, in schizophrenia. Together, they show that genetic factors beyond common inherited mutations can increase risk for schizophrenia. They also point toward biological mechanisms that may be important in schizophrenia, autism and intellectual disability.
Spontaneous shifts:
O’Donovan and his team looked at the genomes of 617 people with schizophrenia and their parents. Looking at the parents’ genes allowed the researchers to determine which mutations in the individual with schizophrenia are spontaneous, or de novo, meaning not inherited from either parent. They compared these de novo mutations with published data from 731 controls.
The rate of de novo mutations is about the same in people with schizophrenia as in controls, the researchers found. But the mutations in people with schizophrenia are significantly more likely to be targets of FMRP, or members of the NMDAR or ARC complexes, two networks of genes associated with synapses. Studies have shown that both networks are important for synaptic plasticity3, 4.
Purcell’s team also found that rare mutations in schizophrenia tend to occur in these gene networks.
“Previously there were dozens of groups of genes one could [consider],” says Dimitri Avramopoulos, assistant professor of psychiatry at Johns Hopkins University in Baltimore, who was not involved in either study. “It’s great to now have more specific subgroups to look at.”
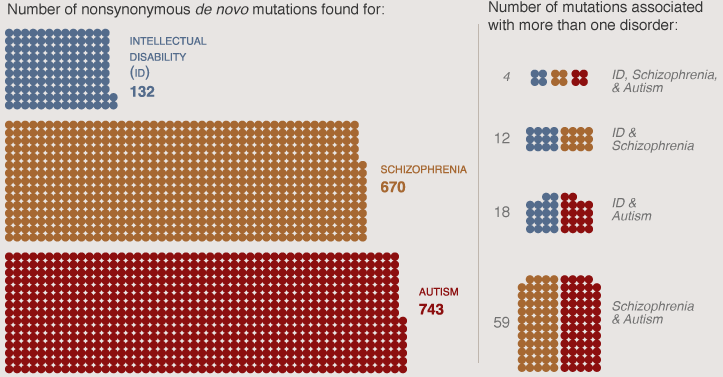
Some of these genes, such as the FMRP targets, have also been found to harbor spontaneous mutations in people with autism or intellectual disability5. Precisely how these mutations influence each of the disorders is still unclear, however.
“The study of mutations alone is unlikely to tell you in a fundamental molecular way in what way [synaptic plasticity] is disrupted,” O’Donovan says. “What we’re doing next is trying to study the impact of the mutations in cells, to see how they adjust the cellular biochemistry or physiology.”
Purcell and his collaborators compared the genomes of 2,536 people with schizophrenia and 2,543 controls. Rather than sequencing every gene from each participant — which would generate a massive amount of data — the researchers focused on about 2,500 genes previously implicated in schizophrenia. They looked in particular for rare variants of genes that, in a sample this size, would be present in only a single individual with schizophrenia.
Overall, they found, rare variants raise schizophrenia risk to a relatively small extent, by about 12 percent on average. But some genes appear to be more important than others. “We were able to identify sets or groupings of genes that had larger effect sizes,” says Pamela Sklar, professor of psychiatry and genetics at Mount Sinai Hospital in New York, who was a collaborator on the study.
This study also found that gene groups involved in neuronal connections and signaling — including the ARC complex genes as well as genes associated with calcium channels — are more likely to be mutated in people with schizophrenia than in controls.
Pinpointing the specific genes that raise risk of the disorder has been difficult, however; because each variant is so rare, it often lacks statistical significance on its own.
“We still can’t point at which individual genes are responsible,” Sklar says. “But we know that as a group, genes from that set confer larger genetic risk.”
When Sklar and her colleagues compared mutations implicated in schizophrenia and autism, they found that only FMRP targets are likely to be involved in both.
“There’s not complete overlap in the genes for the two disorders,” Sklar says. “There’s plenty of room for shared disease effect and individual disease effect.”
The study underscores just how genetically heterogeneous schizophrenia is, adds McClellan. “It isn’t surprising when you’re sequencing 100 individuals that there’s heterogeneity,” he says. “But now you’re sequencing 2,500 and you still have enormous heterogeneity. No single gene by itself explains very many cases.”
References:
1: Fromer M. et al. Nature Epub ahead of print (2014) PubMed
2: Purcell S.M. et al. Nature Epub ahead of print (2014) PubMed
3: Malenka R.C. et al. Trends Neurosci. 16, 521-527 (1993) Abstract
4: Korb E. et al. Trends Neurosci. 34, 591-598 (2011) PubMed
5: Sullivan P.F. et al. Nature Rev. Genet. 13, 537-551 (2012) PubMed
1. Neale, B.M. et al. Nature 485, 242-245 (2012) PubMed
2. O’Roak, B.J. et al. Nature 485, 246-250 (2012) PubMed
3. Sanders, S.J. et al. Nature 485, 237-241 (2012) PubMed
4. Iossifov, I. et al. Neuron 74, 285-299 (2012) PubMed
5. Rauch, A. et al. Lancet 380, 1674-1682 (2012) PubMed
6. deLigt, J. et al. N. Engl. J. Med. 367, 1921-1929 (2012) PubMed
7. Girard, S.L. et al. Nature Genet. 43, 860-863 (2011) PubMed
8. Xu, B. et al. Nature Genet. 44, 1365-1369 (2012) PubMed
9. Gulsuner, S. et al. Cell 154, 518-529 (2013) PubMed
By joining the discussion, you agree to our privacy policy.